Using R to evaluate DGE for differential expression
Moderated statistical tests for assessing differences in tag abundance. (2007). Robinson MD, Smyth GK. Bioinformatics. 23(21):2881-7. PMID: 17881408
The authors of this paper develop a statistical test using a negative binomial distribution to model noise in the data. They release R code (available at bioinf.wehi.edu.au/resources/) to support their treatment, but it is uncommented. The windows version installs from their zip file, but I was unable to get the linux version to build.
Here I apply the code and examine the results in a sample data set.
At the website above you can find a tar archive called: msage_0.7.tar.gz. I can't get the package to install in R, so I simply unpack it and use the .R files directly. In addition, the code calls a code block (kepler.R) which is not included in the distribution. However, it is available here: http://dulci.biostat.duke.edu/sage/sage_analysis_code.r
Incidentally, the block of code: kepler.R is not actually required for the Robinson-Smyth algorithm. It is only called for the comparison of their algorithm to Kepler's algorithm quoted in the paper.
Basically there are two files of R code in the msage package: fitNBP.R and msage.R that can be sourced. The file msage.R has a source statement inside: source("kepler.R"), however this code block is not included. To get the code to function, download sage_analysis.r, and rename it kepler.R (written by Thomas Kepler at Duke). It contains a function called: glm.poisson.disp() that is called by msage.R.
Since I can't get the package to install, I simply place the .R files in my working directory (along with kepler.R), and source them:
# set working directory and load library functions
setwd("E:/cry1/Solexa/Smyth")
source("fitNBP.R")
source("msage.R")
This doesn't strike me as the optimal way to proceed, as you end up with a messy workspace (71 items in your environment), but I don't know a better way.
Assemble some data
There is a function in msage which can assemble data from individual tab-delimited tag files of the form: tag, count. It's called: read.sage() and takes an array of file names to read. It returns a list with two elements: a data matrix in the first position, and an array of library sizes as the second element. The input files simply need two columns, lists of tags (or gene names) and the associated counts separated by a tab character. They do not have to be normalized to tpm.
Assuming you had a directory of tag results files you could read them in as follows:
# read mydir to get a list of file names with .txt extensions
files <- dir("mydir", "txt", full.n=T)
# read in files of my choosing
dge <- read.sage(files[c(3,4,7,8)])
dge will now be a list containing a data matrix with counts for all the tags from all the files that were read, and an array with the total counts found in each file (also known as the size of each library).
However I already have my data in a table.
wt1 wt2 mut1 mut2 NM_008665 3347 2740 3185 2842 CF805324 3336 2897 3500 2838 NM_030248 3252 2754 3231 2604 BC093504 3241 3179 3482 2943 AK043166 3157 7257 55 8837 NM_133703 3153 3196 2839 3588 NM_198419 3131 2787 2631 3576 BX511437 2893 2017 2429 2059 NM_028110 2844 2611 2279 2237 BF470700 2830 2629 3343 2333 AK028146 2681 2464 2104 2501 ...
Once we have some data we need to make a list to hand to the first function. The list contains the data matrix, the sample groupings, and the library sizes. You may want to filter your data to only contain rows of the matrix that have reasonable tag counts. A set of columns with only 1 count in half of them won't be very meaningful.
My data is in a table called gex.all (if it's a data frame you can reference it as.matrix as I do below). I need to find the size of each library (aka the sum of each column), and I want to filter it a little:
lib.size <- colSums(gex.all) # create a filter for really low numbers of reads k <- rowSums(gex.all) > 4 # create a list structure cws <- list(data=as.matrix(gex.all[k,]), groups=c(1,1,2,2), lib.size=lib.size) # run the first function from msage # this takes a while alpha <- alpha.approxeb(cws) # this takes a while ms <- msage(cws, alpha=alpha$alpha) adj.p <- p.adjust(ms$exact, "fdr") k <- (adj.p < .05) # create table with some results all <- cbind(cws$data,adj.p,sign(rowSums(cws$data[,1:2]) - rowSums(cws$data[,3:4]))) # save results to disk write.table(all[k,], "all.csv", sep=",", col.names=NA)
The functions: alpha.approxeb() and msage() took a combined time of 36 hours to run on a data set with 18000 rows.
Plotted below is a snapshot of the results. Spots are labeled with a color gradient based on p-value. Red spots have p-values of 0.01 or less.
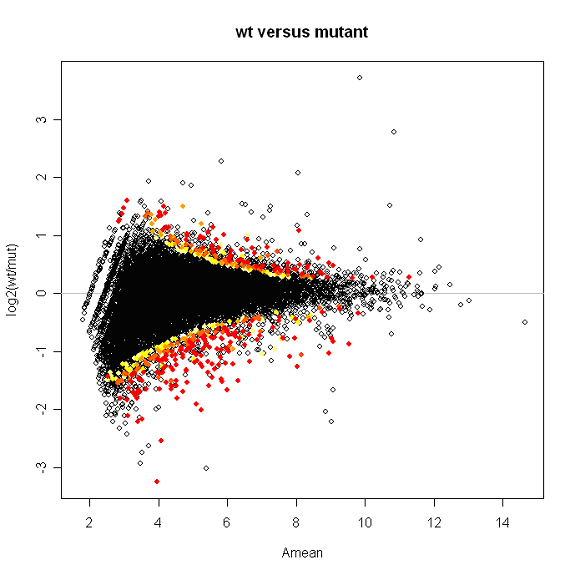
Kepler Sage Analysis
The data can also be analyzed by putting a wrapper around Kepler's algorithm, as described in.
Identifying differential expression in multiple SAGE libraries: an overdispersed log-linear model approach. (2005). Lu J, Tomfohr JK, Kepler TB. BMC Bioinformatics. 6:165. PMID: 15987513
Using the code referenced above (sage_analysis_code.r), for a data matrix containing transcripts per million
# for data matrix mgex
results <- c()
for(i in 1:nrow(mgex)){
tag.count <- as.numeric(mgex[i,])
#fit the overdispersed log-linear model
fit <- glm(tag.count~offset(log(lib.size))+group,family=poisson(link=log));
fit.ps <- glm.poisson.disp(fit,verbose=FALSE);
phi <- fit.ps$dispersion;
t.value <- summary(fit.ps)$coefficients[2,'z value'];
p.value <- 2*pt(-abs(t.value),fit.ps$df.residual);
results <- rbind(results, c(tag.count, t.value, phi, p.value))
# spit out some output so I know things are happening
if( ! i %% 100 ){
cat("row number ", i, "\n")
flush.console()
}
}
rownames(results) <- rownames(mgex)
colnames(results) <- c(colnames(mgex), "t", "phi", "p")
write.table(results, file="kepler_sage_results.txt", sep=",", col.names=NA)
CategoryR ChrisSeidel
