solexa ChIP SEQ data in R
This is an example workflow for analysis of solexa ChIP SEQ data in R.
When processing solexa data for chip SEQ analysis, many of us have our own scripts that have been written in Perl or Python to do things like gnerate BED or WIG files, create IP/wce ratios, etc. However, these scripts take a long time to run, can use vast amounts of memory (which may require that you use specific hardware), and writing custom scripts for specific tasks is very inefficient for exploring data. On the other hand, R is a great environment for exploring data, and packages have been developed for for analysis of solexa data.
Some of the packages for analysis of solexa data are under active development, and may not be ready for primetime. However, in this tutorial I lay out how to read in a data set, and convert it to something which takes a relatively small memory footprint, such that you can manipulate it and explore it with ease.
The only problem with using R for chip SEQ analysis, is that you have to traverse various objects that are at first confusing and unfamiliar. However, once you get used to them, they can be very powerful.
Here is an overview of some of the objects one goes through to analyze chip SEQ data in R:
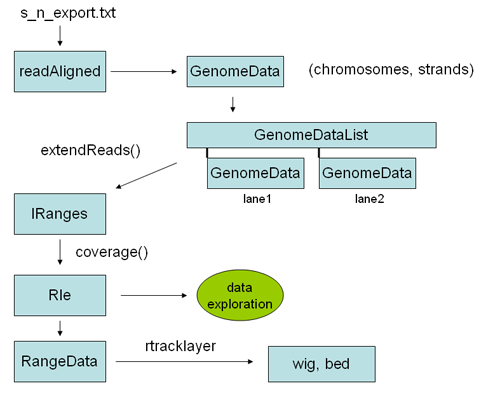
The first step is reading the data into an alignedReads object. This is an object from the ShortRead library, and contains all the relevant solexa data, including sequences and quality scores. To perform genomic analysis on Chip SEQ data most of this information is un-neccesary, and all you really need are the chromosomal locations, including strand, of the alignments. This information can be encoded into a GenomeData object, which is the kind of object required by some of the chipseq library functions. To convert an alignedReads object to a genome data object we can simply coerce is using "as" (see example below). A genomeData object is a list of chromosomes, each with two sub lists of integers for each strand. Thus the GenomeData object encodes the genomic locations, by strand, for every read of interest on every chromosome. Once you have the data in this form, it is typically very small - perhaps 1/100th of the original size of the solexa result file. You can save it as an R object, and copy it to your flash drive for analysis at the cafe.
The basic idea is to go from sequence reads in export format (i.e. from Eland files of the form: s_n_export.txt) to a track you can see in the genome browser.
library(ShortRead)
library(chipseq)
# create a path to the location of your export files
spath <- "/n/analysis/cws42/42TH9AAXX"
# create some filters
nfilt <- nFilter()
# you can use a regular expression to grab only reads
# which align to a set of chromosomes
cfilt <- chromosomeFilter("Drosophila_melanogaster.chr.[234|Het|L|R|X]+.fa")
# quality filter
qfilt <- alignDataFilter(expression(filtering == "Y"))
# combine filters together into one
filt <- compose(nfilt, cfilt, qfilt)
# Read in a lane of data
aln <- readAligned(spath, pattern="s_1_export.txt", type="SolexaExport", filter=filt)
# convert to GenomeData object
aln.gd <- as(aln,"GenomeData")
If you save the code to a file, say export2GD.r, you can execute it on the command line as follows:
cat export2GD.r | R --vanilla
